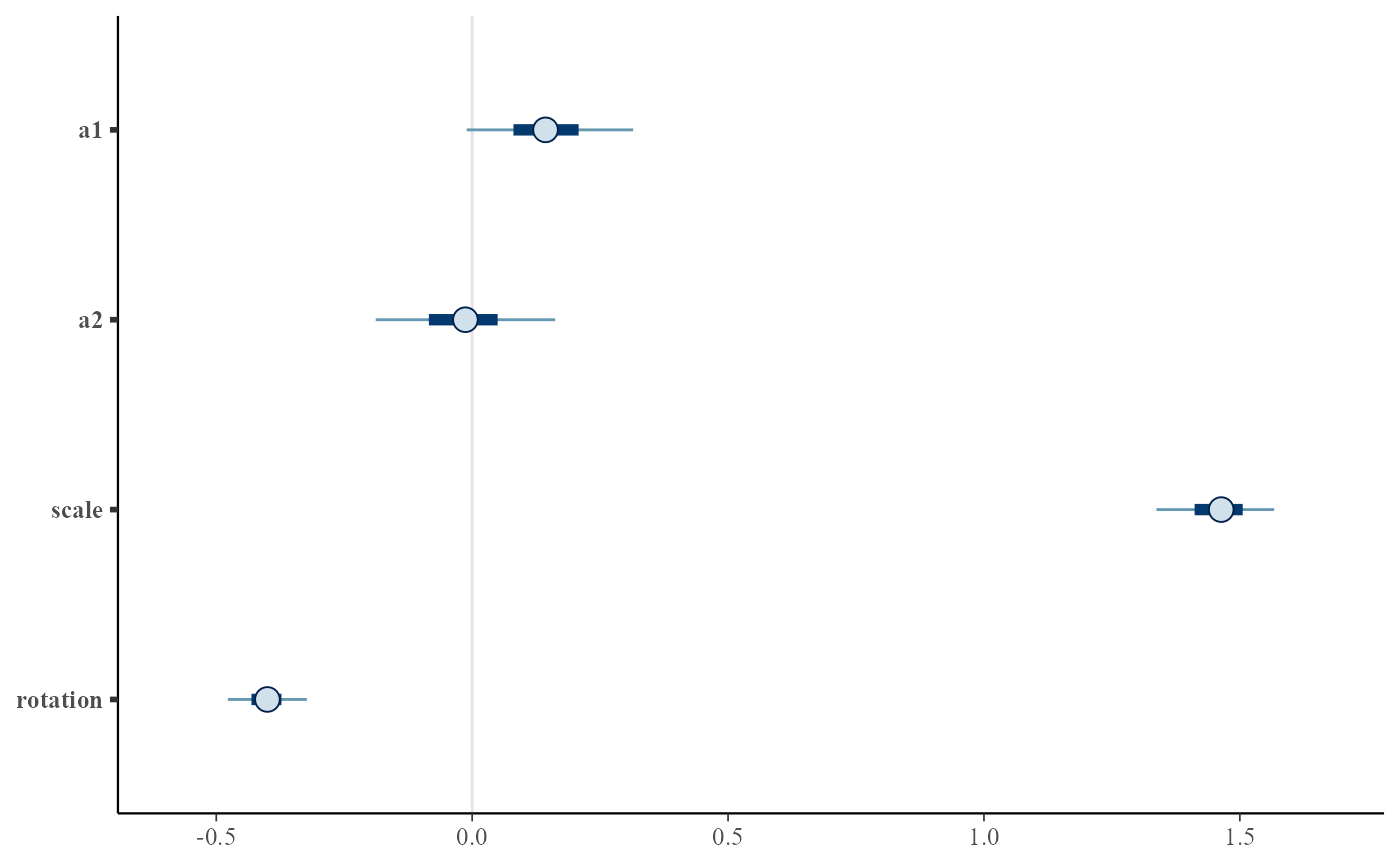
Posterior interval plots for key parameters. Uses bayesplot::mcmc_intervals.
Source:R/plot.R
plot.tridim_transformation.RdPosterior interval plots for key parameters. Uses bayesplot::mcmc_intervals.
Usage
# S3 method for tridim_transformation
plot(x, convert_euclidean = FALSE, ...)Arguments
- x
A tridim_transformation object
- convert_euclidean
Whether to convert matrix coefficients to scale(phi) and rotation(theta). Defaults to
FALSE.- ...
Extra parameters to be passed to
bayesplot::mcmc_intervals()
Value
A ggplot object produced by bayesplot::mcmc_intervals()
Examples
euc2 <- fit_transformation(depV1+depV2~indepV1+indepV2,
data = NakayaData,
transformation = 'euclidean')
#>
#> SAMPLING FOR MODEL 'tridim_transformation' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 0 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.106 seconds (Warm-up)
#> Chain 1: 0.113 seconds (Sampling)
#> Chain 1: 0.219 seconds (Total)
#> Chain 1:
plot(euc2)
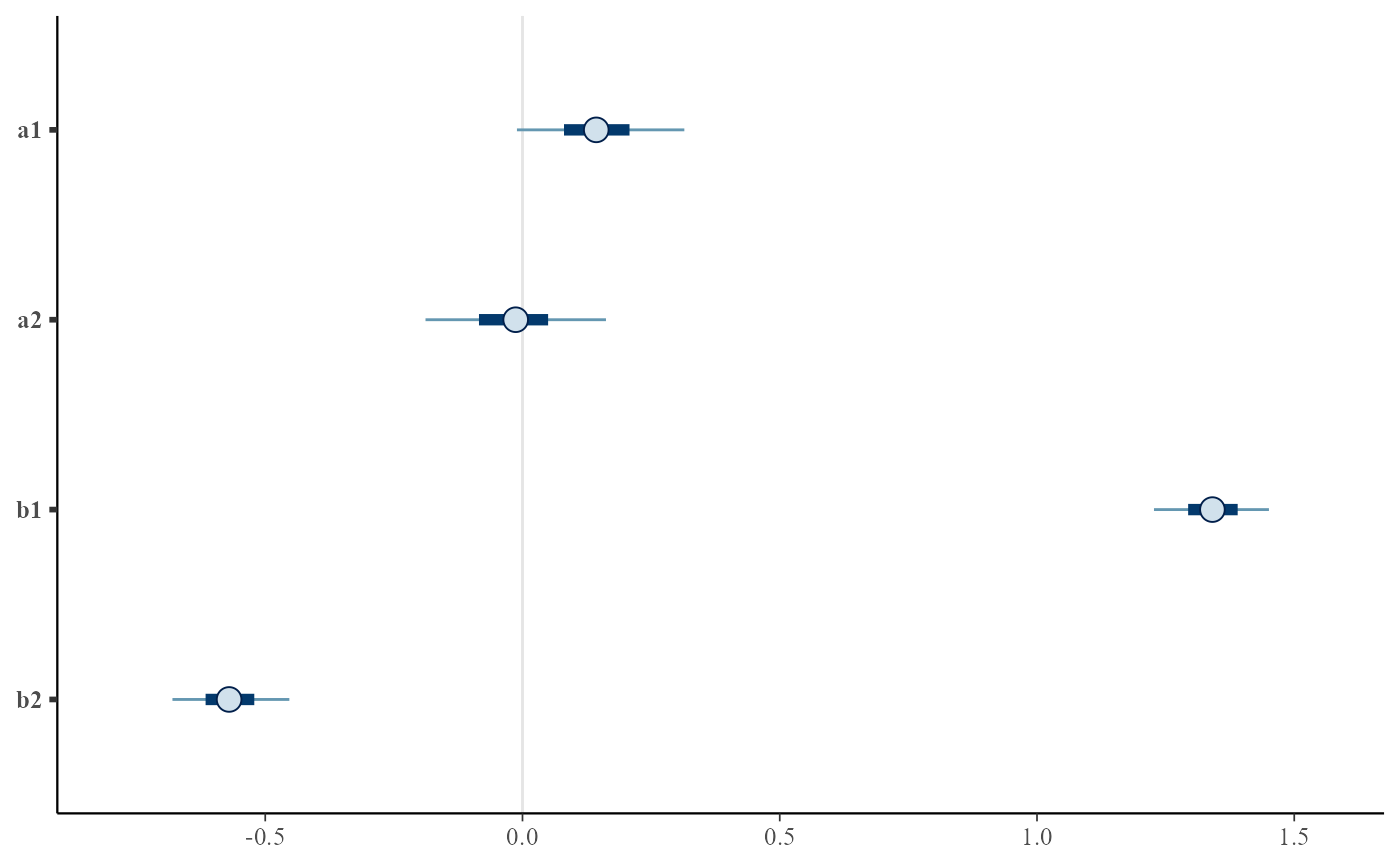 # same but for converted coefficients
plot(euc2, convert_euclidean=TRUE)
# same but for converted coefficients
plot(euc2, convert_euclidean=TRUE)